Functional organisation of the eukaryotic genome
Nucleosomes are the structural units of eukaryotic chromatin. The vast majority of the genome is tightly associated with nucleosomes, which modulate all genomic processes through their interaction with regulatory complexes and through the information encoded in epigenetic modifications of histones. Consistent with these specialised functions, most nucleosomes in the genome occupy precise positions relative to the DNA sequence (Figure 1).
1. Most promoters colocalise with short nucleosome-free regions (NDRs) where transcription initiates (Figure 1). The aim of this line of research is to determine how NDRs manage to exclude nucleosomes through the combined action of remodelers, transcription factors, the mechanical properties of DNA and the +1 nucleosome.
2. We are studying how chromatin remodelers regulate the dynamics of the nucleosome pattern. We use a degron-based approach that allows the transient and reversible inactivation of remodelers to assess how modifying the position of nucleosomes in vivo affects the expression pattern of genes involved in different physiological conditions (Figure 2).
3. We are also interested in how chromatin organisation and transcriptional patterns are regenerated after their physical disruption during double-strand break repair using an experimental approach based on genomic sequencing that we have developed recently (Figure 3) (Ramos et al. 2022).
Given the universality of DNA-histone interactions and the conserved regulatory role of chromatin in fundamental genomic processes, we use the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae as model systems for biochemical, genetic, genomic, bioinformatic and computational approaches. We exploit the feasibility of genome manipulation in these yeasts to establish cause-effect relationships between genome manipulation, chromatin organisation and gene expression at a level of resolution that is difficult to achieve in animal cells.
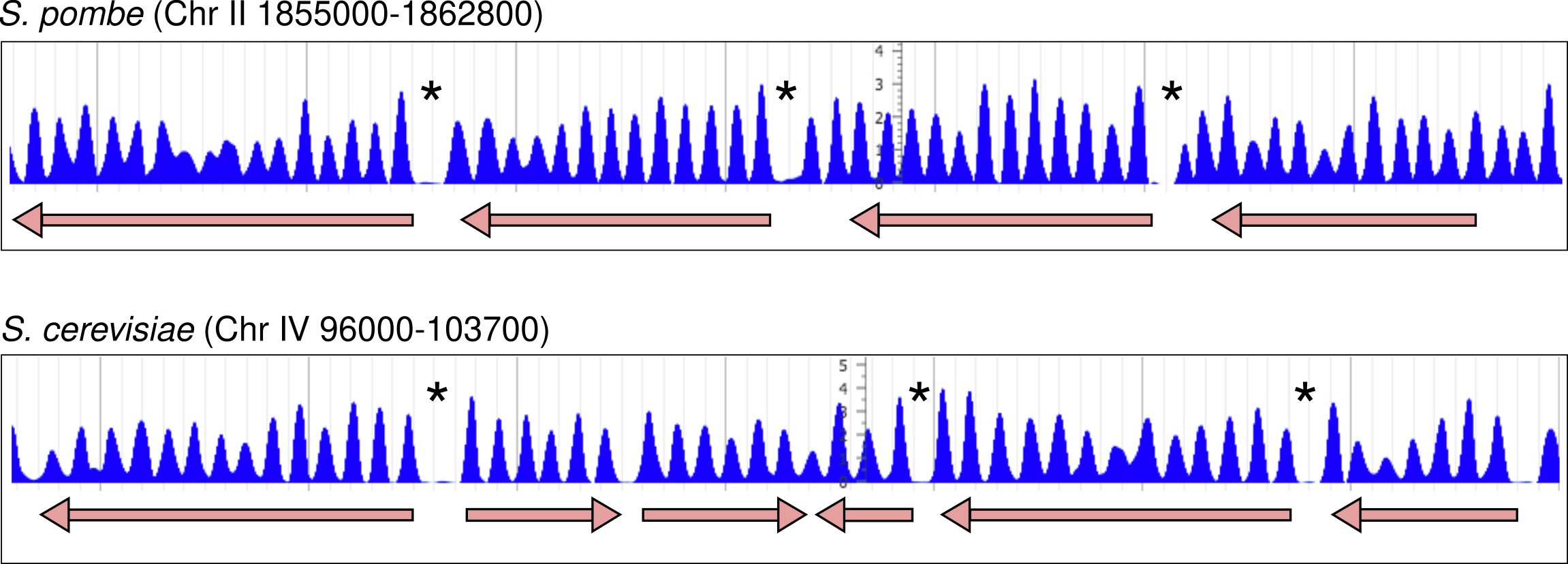
Figure 1. Nucleosomal organisation in S. pombe and S. cerevisiae.
Distribution of nucleosomes along two genomic regions at the indicated genomic positions in chromosomes II and IV. Genes and direction of transcription are indicated by arrows. Asteriks indicate the location of nucleosome-free regions (NDR) at the 5’ end of most genes.
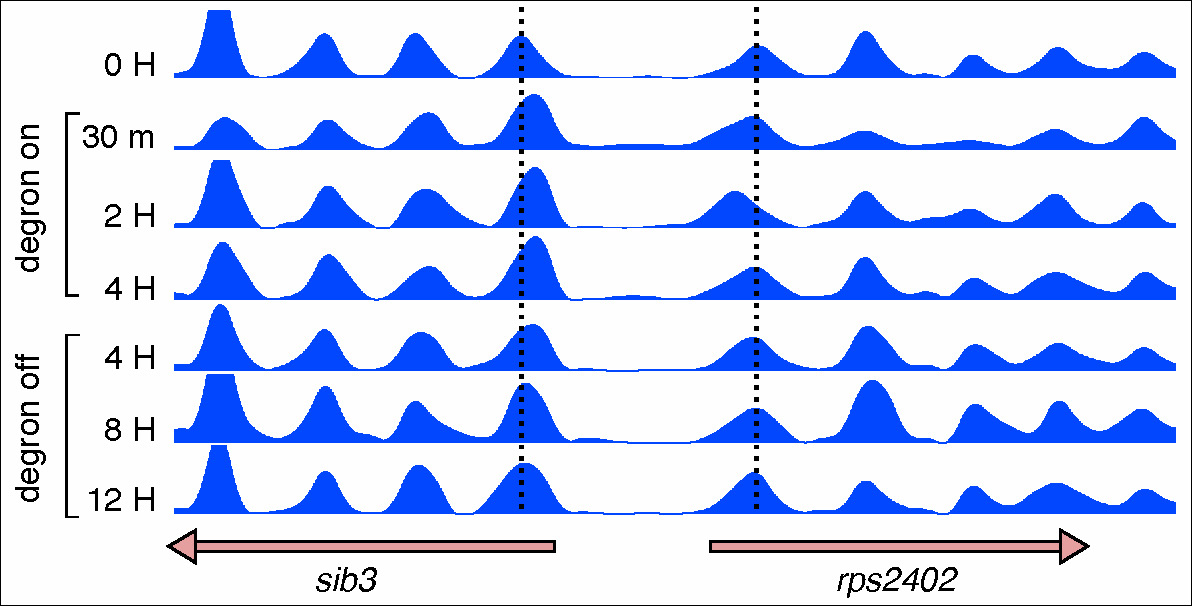
Figure 2. Reversible loss of nucleosome positioning upon remodeler inactivation.
Nucleosome occupancy maps of a S. pombe strain carrying a degron-tagged Snf21 catalytic subunit of the RSC remodeler. Vertical dashed lines indicate the dyad position of +1 nucleosomes of the divergent sib3 and rps2402 genes. These nucleosomes are displaced towards the NDR 30 minutes after degron induction and return to their wild-type position 8-12 hours after degron switch-off.
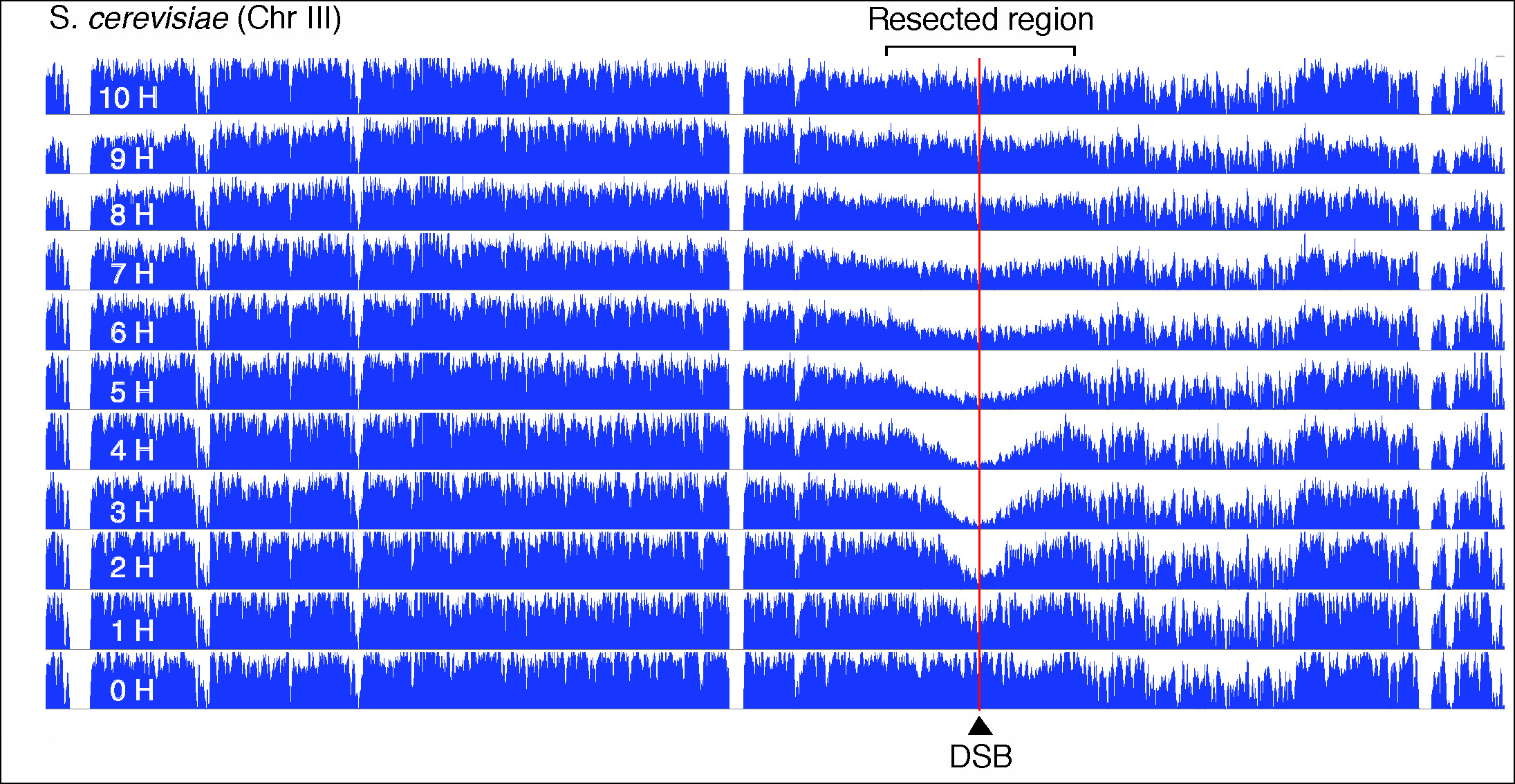
Figure 3. Visualisation of DSB repair of S. cerevisiae chromosome III by genomic sequencing.
The figure shows the coverage along the entire chromosome III of S. cerevisiae at the indicated times after induction of a double-strand break (DSB) at position 200753. The extension of the resection is visualised by the decrease and recovery of coverage in the region flanking the DSB (red line).

Miembros del grupo
| Francisco Antequera | Profesor de Investigación (CSIC) |
|---|---|
| Laura Durán | Postdoctoral |
| Alicia García | Postdoctoral |
| Loreto Megido | Predoctoral |
| Laura Rodríguez | Predoctoral |
| Mar Sánchez | Técnico Titulada Superior |
| Rodrigo Santamaría | Profesor Titular (USAL) |
| Isabel Vaquero | Predoctoral |
Contacto
| Francisco Antequera |
cpg@usal.es 923294915 Laboratorio 2.5 |
|---|
Publicaciones recientes
| García A, Durán L, Sánchez M, González S, Santamaría R, Antequera F. (2024)
Asymmetrical nucleosomal DNA signatures regulate transcriptional directionality. Cell Reports 43: 113605 Doi: 10.1016/j.celrep.2023.113605 |
| Schindler D, Walker R; Jiang S, Brooks A, Wang Y, Mueller C, Cockram C, Luo Y, García A, Schraivogel D, Mozziconacci J, Pena N, Assari M, Sanchez M, Zhao Y, Ballerini A, Blount B, Cai J, Ogunlana L, Liu W, Joensson K, Abramczyk D, Garcia-Ruiz E, Turowski T, Swidah R, Ellis T, Pan T, Antequera F, Shen Y, Nieduszynski C, Koszul R, Dai J, Steinmetz L, Boeke J, Cai Y. (2023)
Design, construction, and functional characterization of a tRNA neochromosome in yeast. Cell 186: 1-17 Doi: 10.1016/j.cell.2023.10.015 |
| Ramos F, Durán L, Sanchez M, Campos A, Hernández-Villamor D, Antequera F*, Clemente-Blanco A*. (2022)
Genome-wide sequencing analysis of Sgs2, Exo1, Rad51, and Srs2 in DNA repair by homologous recombination. Cell Reports 38: 1-17 (*Co-corresponding authors) Doi: 10.1016/j.celrep.2021.110201 |
| García J, Lazar-Stefanita L, Gutierrez-Escribano P, Thierry A, García A, González S, Sánchez M , Jarmuz A, Montoya A, Dore M, Kramer H, Mehdi Karimi M, Antequera F, Koszul R, Aragon L. (2019)
FACT mediates cohesin function on chromatin. Nature Struct. Mol. Biol. 26: 970-979 Doi: 10.1038/s41594-019-0307-x |
| Santamaría R, Therón R, Durán L, García A, González S, Sánchez M and Antequera F (2018)
Genome-wide search of nucleosome patterns using visual analytics Bioinformatics 1-8 Doi: 10.1093/bioinformatics/bty971 |
| González S, García A, Vázquez E, Serrano R, Sánchez M, Quintales L and Antequera F (2016)
Nucleosomal signatures impose nucleosome positioning in coding and non-coding sequences in the genome Genome Research 26: 1532-1543 Doi: 10.1101/gr.207241.116 |
| Quintales L, Soriano I, Vázquez E, Segurado M and Antequera F (2015)
A species-specific nucleosomal signature defines a periodic distribution of amino acids in proteins Open Biology 5: 140218 Doi: 10.1098/rsob.140218 |
| Castel E, Ren J, Bhattacharjee S, Chang A, Sánchez M, Valbuena A, Antequera F & Martienssen R (2014)
Dicer promotes transcription termination at sites of replication stress to maintain genome stability Cell 159: 572-583 Doi: 10.1016/j.cell.2014.09.031 |







