From Genes to Chromosomes - The Chromosome Biology Lab
Eukaryotic genomes are packed into chromatin so that long molecules of DNA, approximately 2 meters in each human cell, fit within a few micrometers of the cell nucleus. The basic unit of chromatin is the nucleosome, which comprises ~147 bp of DNA wrapped around a histone octamer.
Nucleosomes prevent the access of DNA to enzymes involved in DNA activities, such as replication, transcription, or DNA repair. Thus, nucleosome organization plays an important role in genome regulation. Nucleosome positions along the genome are determined by several factors, including the underlying DNA base pair composition, histone post-translational modifications, transcription factors and the action of chromatin-remodeling complexes, specialized protein machines that utilize ATP hydrolysis to mobilize nucleosomes. While the relative contribution of each of these factors and their interdependency on one another is still not fully understood, plenty genetic and biochemical evidence place chromatin remodelers at the core of the regulation of the primary structure of chromatin.
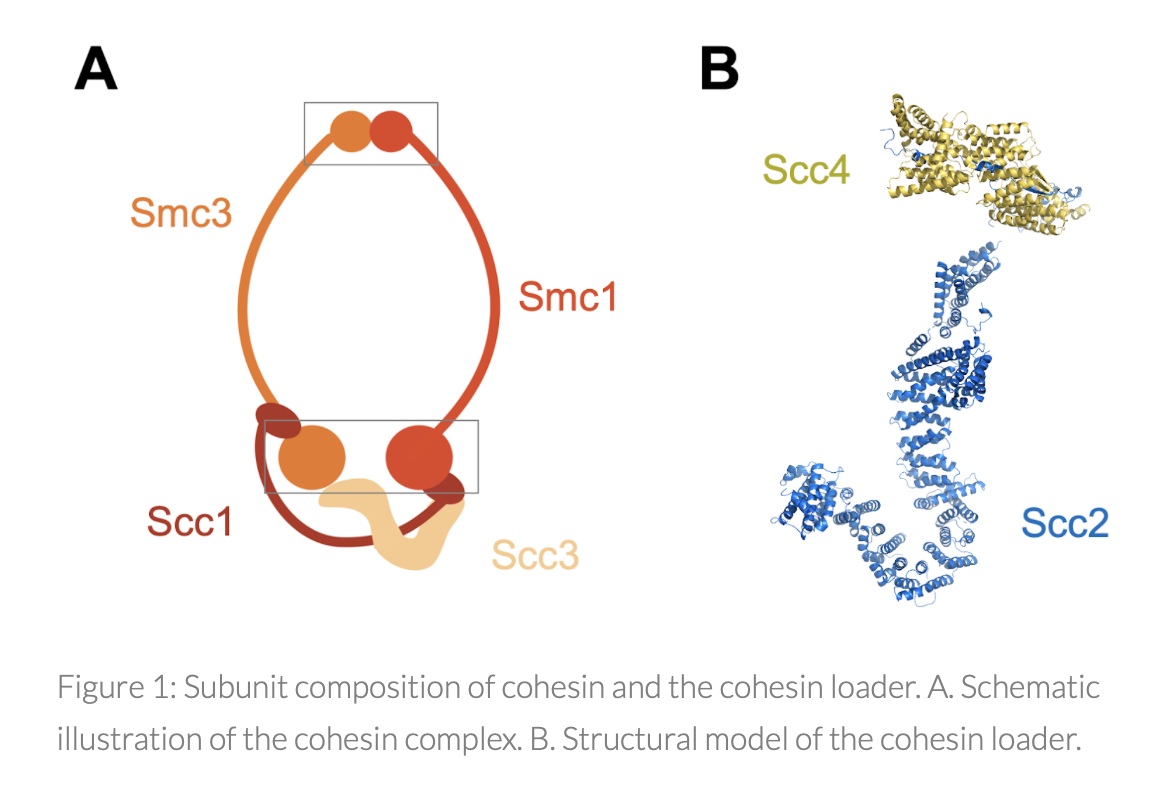
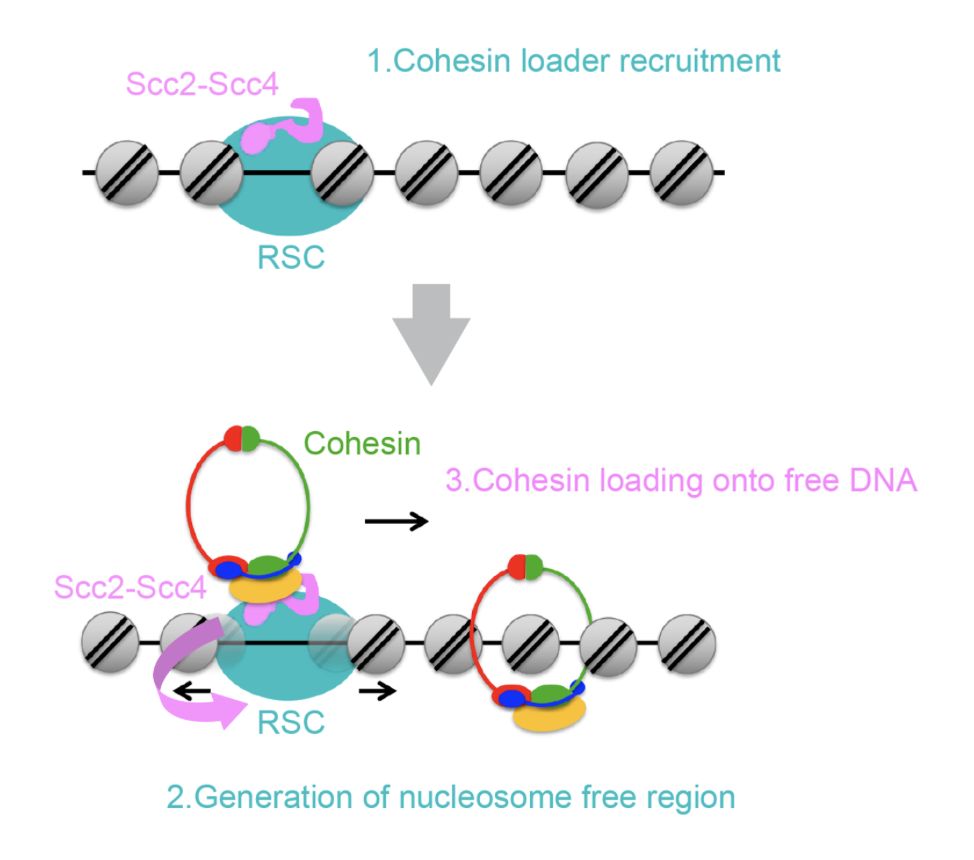
The nucleosome fiber is subsequently folded through DNA looping by the action of the SMC ‘Structural Maintenance of Chromosomes’ proteins, conserved ring-shaped complexes fueled by ATP hydrolysis that encircle DNA and organize the genome by building loops. Member of the SMC family, the cohesin complex embraces sister chromatids together from DNA replication until metaphase in a process known as sister chromatid cohesion, which is essential for accurate chromosome segregation during mitosis. Besides sister chromatid cohesion, cohesin has crucial roles in other structural and functional aspects of chromosomes, including genome organization during interphase, gene transcription and DNA repair. The loading of cohesin onto chromosomes requires a second protein complex Scc2-Scc4 known as the cohesin loader.
During our research we have discovered that nucleosomes inhibit the loading of cohesin onto chromosomes and that a chromatin remodeler, RSC, plays a fundamental role in cohesin loading onto chromatin. On the other hand, we have discovered that the cohesin loader stimulates RSC remodeling activity, contributing to promoter nucleosome clearance, with important potential implications in the regulation of gene expression.
Our research explores the possibility that local chromatin structure regulates higher order chromatin conformations by regulating SMC complexes, the main regulators of the DNA folding. In addition, SMC proteins might feed back onto the regulation of nucleosome positioning by regulating chromatin remodelers.

Miembros del grupo
| Sofía Muñoz | Group Leader |
|---|
Contacto
| Sofía Muñoz |
sofiamf@usal.es +34 923294900 Laboratorio 1.3 |
|---|
Publicaciones recientes
| Sofía Muñoz; Andrew Jones; Céline Bouchoux; Tegan Gilmore; Harshil Patel; Frank Uhlmann (2022)
Functional crosstalk between the cohesin loader and chromatin remodelers . Nature Communications |
| Mark Mattingly; Chris Seidel; Sofía Muñoz; Yan Hao, Ying Zhang; Zhihui Wen; Laurence Florens; Frank Uhlmann; Jennifer L. Gerton (2022)
Mediator recruits the cohesin loader Scc2 to RNA Pol II transcribed genes and promotes sister chromatid cohesion Current Biology |
| Sofía Muñoz; Francesca Passarelli; Frank Uhlmann (2020)
Conserved roles of chromatin remodellers in cohesin loading onto chromatin Current Genetics |
| Sofía Muñoz ; Masashi Minamino; Corella S. Casas-Delucchi; Harshil Patel; Frank Uhlmann (2019)
A Role for Chromatin Remodeling in Cohesin Loading onto Chromosomes Molecular Cell |
Proyectos de investigación
| PID2023-149921NA-I00 |
| MICINN: RYC2022-037334-I |







