Quantitative Biology
Our research centers on exploring how evolutionary biology may help us make sense, predict, and engineer microbial communities. To this end, we employ a combination of laboratory experiments, computer simulations, and mathematical modeling. Our laboratory is highly interdisciplinary and quantitative in nature, and our students and postdocs typically combine theory and experiment in their projects, acquiring a broad range of technical skills across the wet-lab / dry-lab divide. They also get exposed to a broad scientific intellectual background in ecology, evolution and systems biology. Our current lines of work are described below.
1) Engineering microbial consortia from the top-down. For millennia, selective breeding has allowed us to artificially select crops and domestic animals. In more recent times, the idea of directed evolution has been extended to engineering biological systems at or below the organismal level: from enzymes and RNA molecules to genetic circuits, metabolic networks, and microbial strains. Can these approaches be also extended to engineering biological systems above the organismal level of organization, such as microbial consortia? Our laboratory is exploring how one may do this successfully (Chang et al 2021, Nature Ecol Evol, Sanchez et al 2021 Ann Rev Biophys, Chang et al 2020 Evolution). We seek to develop empirical methods, grounded on ecological and evolutionary theory, that may be then adapted by applied biotechnologists in their particular settings. To fund the work in this research line, we have been awarded a project from the Spanish Agencia Estatal de Investigacion, MICROBREED: Extending evolutionary engineering to microbial consortia.
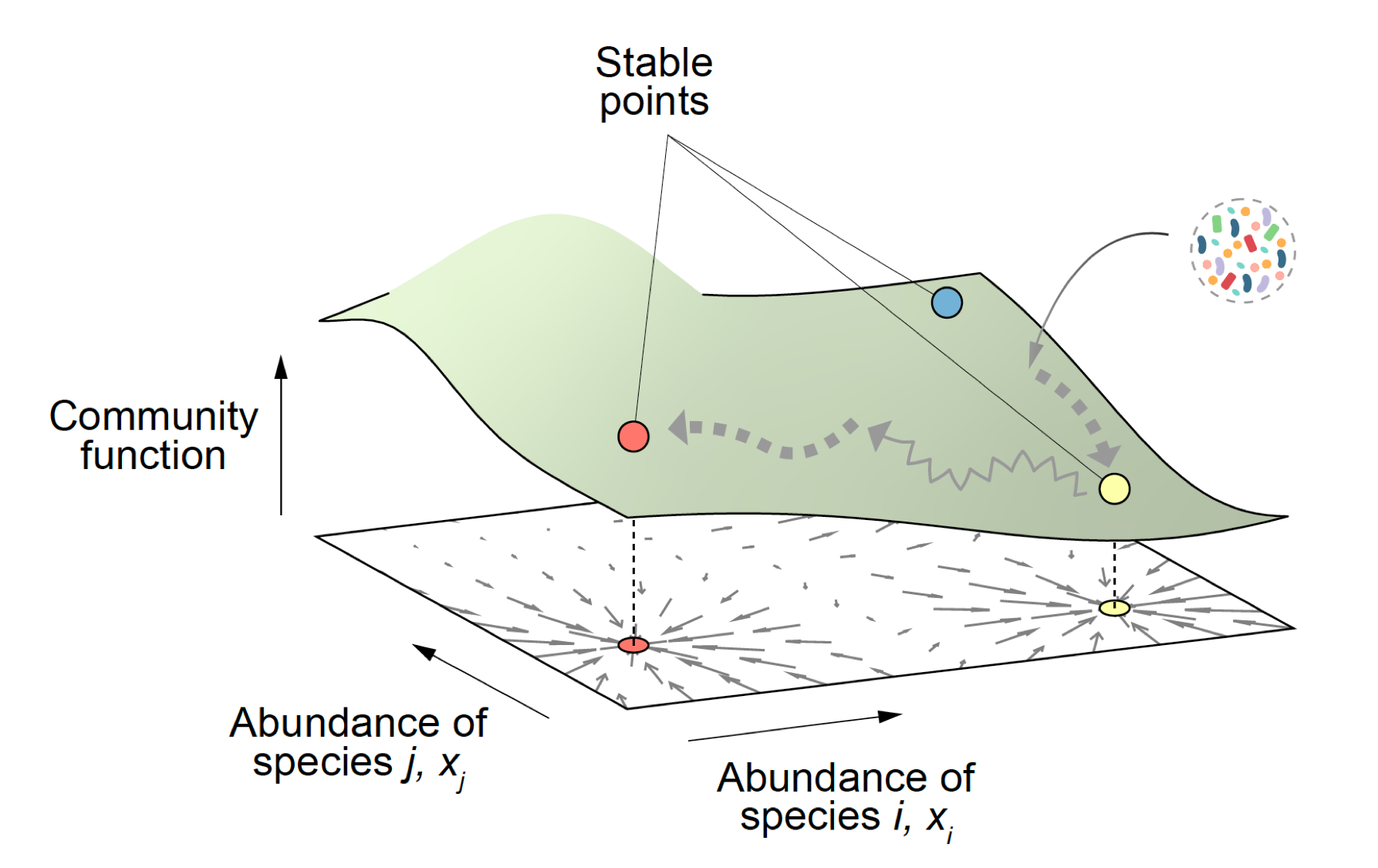
2) Predictively linking microbial community composition and community function. Microbial communities provide us with countless ecological services that are essential for the health of the biosphere and to preserve life as we know it. They also carry out a vast and growing number of important functions in biotechnology, from food production to the synthesis of biofuels and other economically critical molecules. These community functions are determined by community composition, i.e. by which genotypes are found in each community and their abundance. If we wish to engineer microbial consortia to optimize the functions they provide, it is key that we predictively and quantitatively link community composition and function. This is a hard problem, that cannot be solved empirically due to the astronomical number of potential consortia one may form even with a modest number of potential candidate genotypes. Microscopic models are exceedingly difficult to build, due to the complex web of interactions that are involved. Our laboratory is exploring whether ideas from the theory of fitness landscapes in genetics might help us solve this problem and produce predictive and quantitative models of community function from its composition. (Sanchez-Gorostiaga et al 2019 PLoS Biol, Lino et al 2021 Nature Comm, Sanchez 2019 Cell Systems, Sanchez et al 2023 Cell Systems, Diaz-Colunga et al 2023 Phil Trans Roy Soc). To fund the work in this research line, we have been awarded an ERC Consolidator Grant.

Group members at the IBFG
| Alvaro Sanchez | Principal Investigator | Belén Suarez | Prof. Contratado Doctor (USAL) | -->
|---|---|
| Magdalena San Roman | Postdoctoral researcher |
| Andrea Arrabal | Technician |
| Belen Benítez Domínguez | PhD student |
| Isabel Quiros | PhD student |
Contact
| Álvaro Sánchez |
alvaro.sanchez@usal.es 923294900 Laboratorio 1.6 |
|---|
Recent publications
| Diaz-Colunga J*, Skwara A*, Vila JCC, Bajic D, & Sanchez A (2024)
Global epistasis and the emeegence of function in microbial consortia. Cell. In press. |
| De la Fuente J*, Diaz-Colunga J*, Sanchez A# & San Millan A#. (2024)
Global epistasis in plasmid-mediated antimicrobial resistance. Molecular Systems Biology. 20: 311-320. |
| Sanchez A, Arrabal A, San Roman M, Diaz-Colunga J. (2024)
The optimization of microbial community functions through rational environmental manipulations. Molecular Microbiology. 10.1111/mmi.15236. |
| Diaz-Colunga, J.#, Sanchez A.#, & Ogbunugafor, C.B.# (2023)
Environmental modulation of global epistasis is governed by effective genetic interactions. Nature Communications. In press. |
| Chang, C-Y. #, Bajic, D. #, Vila, J.C.C., Estrela, S. & Sanchez A. # (2023)
Emergent coexistence in multispecies microbial communities. Science 381(6655) 343-348. |
| Skwara A., Gowda K., Yousef M., Diaz-Colunga, J., Raman A., Sanchez A., Tikhonov M. & Kuehn S. (2023)
Statistically learning the functional landscape of microbial communities. Nature Ecology & Evolution 7: 1823-1833. |
| Ruiz, J., De Celis, M., Diaz-Colunga, J., Vila J.C.C., Benitez-Dominguez, B., Vicente J., Santos, A., Sanchez A., & Belda, I. (2023)
Predictability of the community-function landscape in wine yeast ecosystems. Molecular Systems Biology e11613 |
| Sanchez A. #, Bajic, D, Diaz-Colunga, J., Skwara, A., Vila, J.C.C. & Kuehn S# (2023)
The community-function landscape of microbial consortia. Cell Systems 14(2) 122-134 |
| Diaz-Colunga, J.*, Skwara, A*., K. Gowda, R. Diaz-Uriarte, M. Tikhonov, Bajic, D. & Sanchez A. # (2023)
Global epistasis on fitness landscapes. Philosophical Transactions of the Royal Society B 378: 20220053 |
| Diaz-Colunga, J.*, Lu, N.*, Sanchez-Gorostiaga, A.*, Cai, H., Chang, C-Y., Goldford, J., Tikhonov, M., Sanchez, A. # (2022)
Top-down and bottom-up cohesiveness in microbial community coalescence.. Proceedings of the National Academy of Sciences 119:6 e2111261119 |
| Estrela S., Vila J.C.C., Lu N., Bajic D., Rebolleda-Gomez M., Chang C-Y., & Sanchez A#(2022)
Functional attractors in microbial community assembly Cell Systems 13-29-42 |
| Bergelson J.*, Kreitman M.*, Petrov D.*, Sanchez A.*, Tikhonov M.* (2021)
Functional biology in its natural context: a search for emergent simplicity. eLife 10:e65948 |
| Chang C-Y.*, Vila J.C.C.*, Bender M., Li R., Mankowski M.C., Bassette M., Borden J., Golfier S., Sanchez P.G., Waymack R., Zhu X., Diaz-Colunga J., Estrela S., Rebolleda-Gomez M. & Sanchez A. # (2021)
Engineering complex communities by directed evolution. Nature Ecology & Evolution 5:1011–1023 |
| Lino F.S.O., Bajic D., Vila J.C.C., Sanchez A. & Sommer M.O.A. (2021)
Complex yeast-bacteria interactions shape the yield of industrial ethanol fermentations. Nature Communications 12:1498 |
| Sanchez A. #, Vila J.C.C., Chang C-Y., Diaz-Colunga J., Estrela S. & Rebolleda-Gomez M. (2021)
Directed evolution of microbial communities. Annual Review of Biophysics 50:323-341 |
| Vila J.C.C, Yang, Y-L, & Sanchez A. #. (2020)
Dissimilarity-Overlap Analysis of replicate enrichment communities. ISME Journal 14:2505-13 |
| Sanchez, A#. (2019)
Defining high-order interactions in synthetic ecology: lessons from physics and quantitative genetics. Cell Systems 9(6): 519-20 |
| Sanchez-Gorostiaga, A.*, Bajic, D.*, Osborne, M.L., Poyatos, J.F., Sanchez, A. # (2019)
High-order interactions distort the functional landscape of microbial consortia. PLoS Biology 17(12): e3000550 |
| Bajic, D.* #, Vila, J.C.C.*, Blount, Z.D., Sanchez, A. # (2018)
On the deformability of an empirical adaptive landscape by microbial evolution. Proceedings of the National Academy of Sciences 115(44):11286-11291 |
| Goldford, J.*, Lu, N.*, Bajic, D., Estrela, S., Sanchez-Gorostiaga, A., Tikhonov, M., Segre, D., Mehta, P. #, Sanchez, A. # (2018)
Emergent simplicity in microbial community assembly. Science 361:469-74 |
Research grants
| MICINN | PID2021- 25478NA-100 |
|---|---|
| ERC-Co | 101088469 |







